Show and edit data
- 1) Locked and unlocked data
- 2) Sorting data
- 3) Edit data under unlocked status
- 4) Set coding or non-coding sequence, and genetic code
- 5) Switch between uppercase and lowercase
- 6) Length and comment of sequence
- 7) Undo
- 8) Save as
- 2) Sorting data
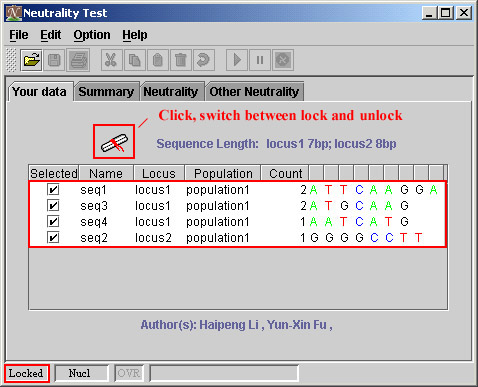
You must understand the locked and unlocked status before editing. It's possible to type a key when you don't know the key is pressed. That might be a disaster if the key is interpreted as an inserted nucleotide. That has never happened under the default configure because your data is locked. You can not edit the sequences under locked status. The following figure shows you the locked data.
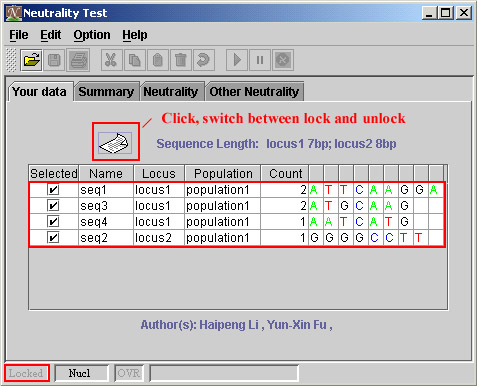
The following figure shows you the unlocked data.
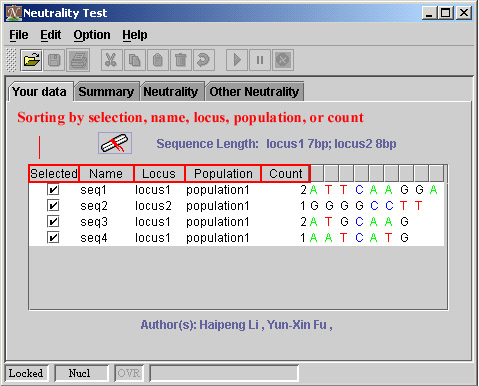
Just click the head of column, then see what happened.
3) Edit data under unlocked status
The software does not provide the aligning function, so you should do the alignment outside, or use highly-similar sequences.
During editing, you can select one sequence, unselect it, rename it, rename the locus, rename the population or species which it belongs to, change the count of sequence (more than or equal to 1), insert or delete a nucleotide. You just need to press "SPACE", or "-" when you want to insert an ins/del. Moreover, you can not input "Q", "H", or "W" as a nucleotide when you are editing sequences. Nothing happens when you type "Q", "H", or "W". The valid input is "A", "a", "T", "t", "C", "c", "G", "g", "-", space key, delete key, and backspace key.
Instruction for selecting and unselecting, change the names and count.
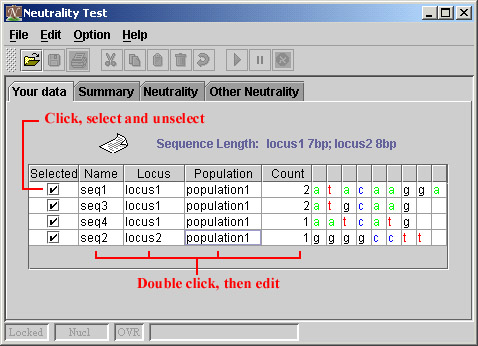
Instruction for editing sequences.
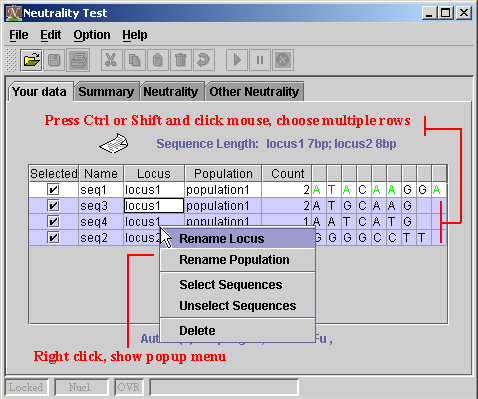
There is an easy way to edit multiple sequences when you need select or unselect them, or rename the name of locus which they belong to, or rename the name of population which they belong to, or delete them.
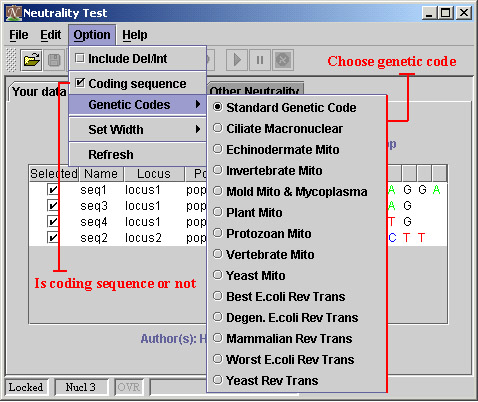
4) Set coding or non-coding sequence, and genetic code
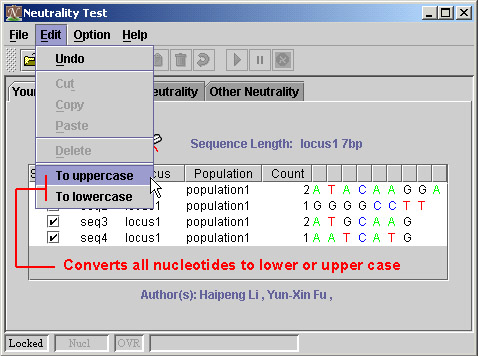
5) Switch between uppercase and lowercase
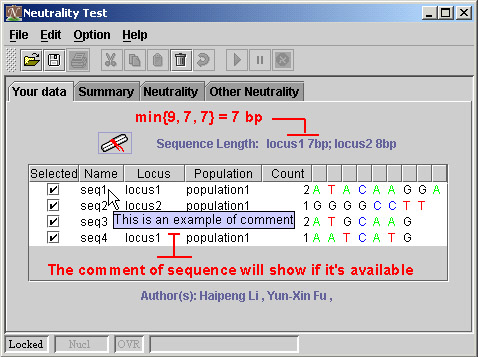
6) Length and comment of sequence
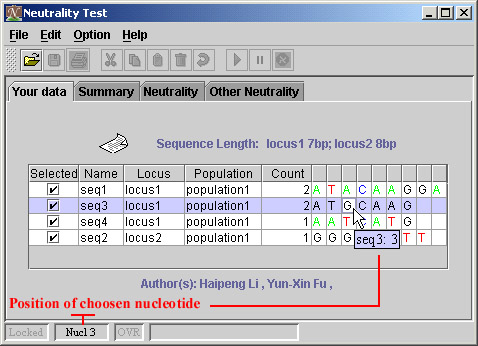
There are three sequences of locus1, the length of them are 9, 7, 7, respectively. Then the length of sequences will be 7, which means the 8-th and 9-th nucleotide in the first sequence will be kicked out the further analysis.
You can define the comment of a sequence, which help you to record some important information on the sequence. The tooltip will show it to you when you put the cursor on name of sequence, and when the comment is available.
You can undo 5 times. Click the undo button on the tool bar, or choose Edit menu, then choose undo item.
You can save your data as different format. However, there is a serious problem when the length of sequences is different. Many formats, such as MEGA and PHYLIP, do not support the "unequal length". In this case, you must edit the saved file before using it as the input file of MEGA or PHYLIP.