The majority of human genes (>95%) are alternatively spliced to produce distinct variants with different functions, the mis-regulation of this process is a common cause for many human diseases including cancer and neurodegenerative diseases. In order to understand the regulation of splicing, we need to solve a set of splicing rules (i.e. splicing code) by collecting the “part lists” of splicing regulatory elements (enhancers or silencers) and further understand how they affect splicing in different contexts. Since introns account for >90% of pre-mRNA, there are a great number of splicing enhancers or silencers in intron need to be identified.
Systematic study of intronic splicing regulatory elements
The regulation of splicing is mostly achieved by RNA cis-elements to recruit protein trans-factors that either enhance or suppress the use of adjacent splice sites. Based on their activities and locations in pre-mRNA, these cis-elemens are classified as exonic splicing enhancers (ESEs), exonic splicing silencers (ESSs), intronic splicing enhancers (ISEs) and intronic splicing silencers (ISSs). Such regulation is analogous to the transcription regulation in which the transcription factor binding to the regulatory cis-elements of DNA and act as enhancers or suppressors of transcription.
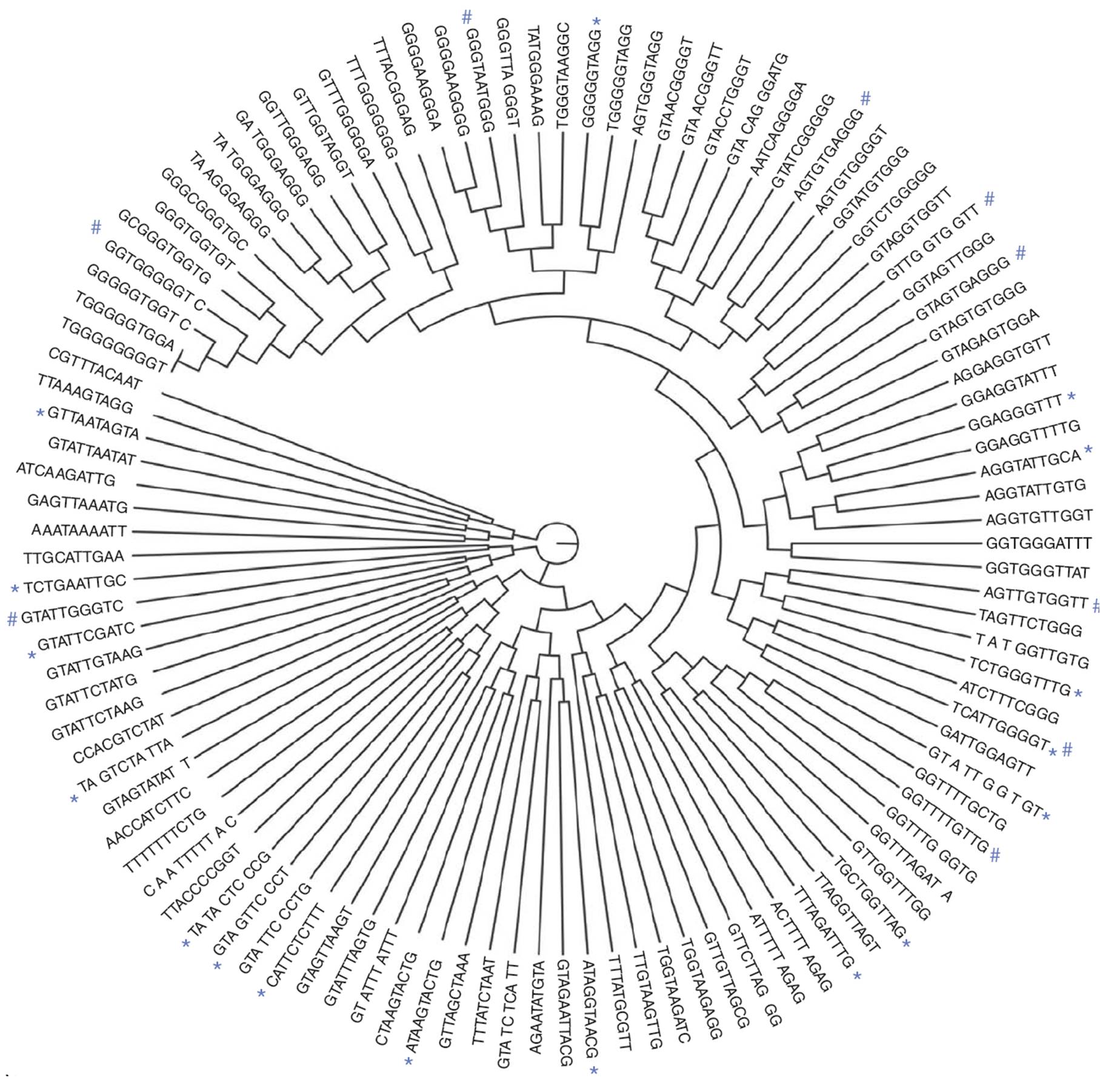
Since most intronic splicing regulatory elements (SREs) are unknown, we seek to identify these cis-elements and study their function systematically. We mainly use a cell based screen to identify novel cis-elements from a random sequence library. In addition, the computational approaches will be used to predict the function of these elements.
We developed new systems to unbiasedly identify intronic splicing enhancers and silencers from random sequences. We identified more than a hundred new intronic splicing enhancers (ISEs) or silencers (ISSs) that were clustered by sequence similarity into six groups of ISEs or ten groups of ISSs. All these elements functioned in another cell type and heterologous introns, and they can either enhance or silence splicing from an exonic context. We will study the mechanism behind their context dependent activities with both computational and experimental approaches. We seek to provide a comprehensive picture of ISS and ISE activities and discovered new models of how a single element can function oppositely depending on its location and binding factors.
Trans-acting splicing factors and their targets
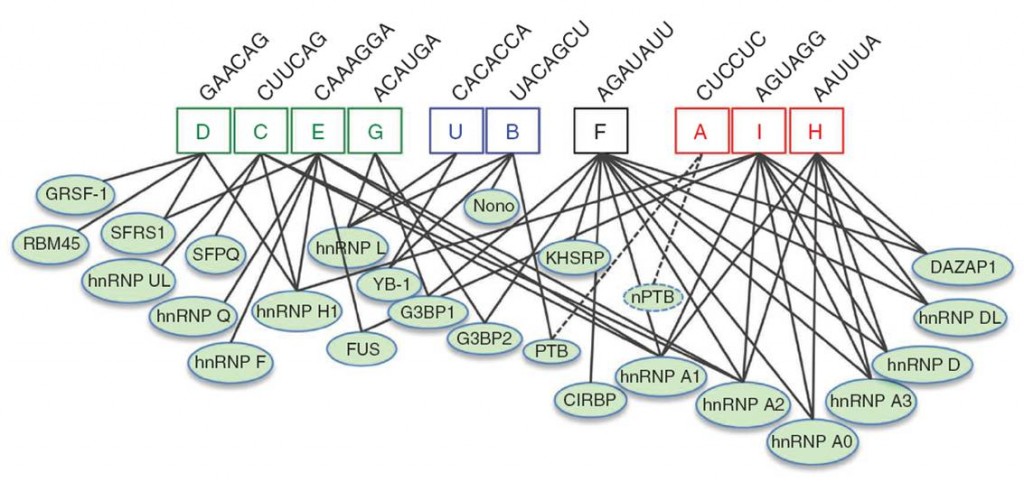
We are also interested in identifying the trans-factors that bind to the regulatory cis-elements. Contrary to earlier model, the interaction between splicing factors and SREs are not one-to-one relationship, rather they form a densely connected interaction network that may provide some regulatory plasticity for splicing. The majority of such factors contain one or more RNA recognition domains and a motif with repetitive sequences, and often function in a modular fashion (i.e. contain a RNA binding module to recognize pre-mRNA target and a functional module to activate or inhibit splicing). We are in the process to study how the interaction network between trans-acting splicing factors and SREs shape the splicing profile inside a cell. This has great therapeutic implication as these splicing factors may serve a drug target to restore the normal splicing. Once we found critical splicing changes in human diseases, we may be able to predict what splicing factors can cause the change and direct new therapeutic method to target these factors.
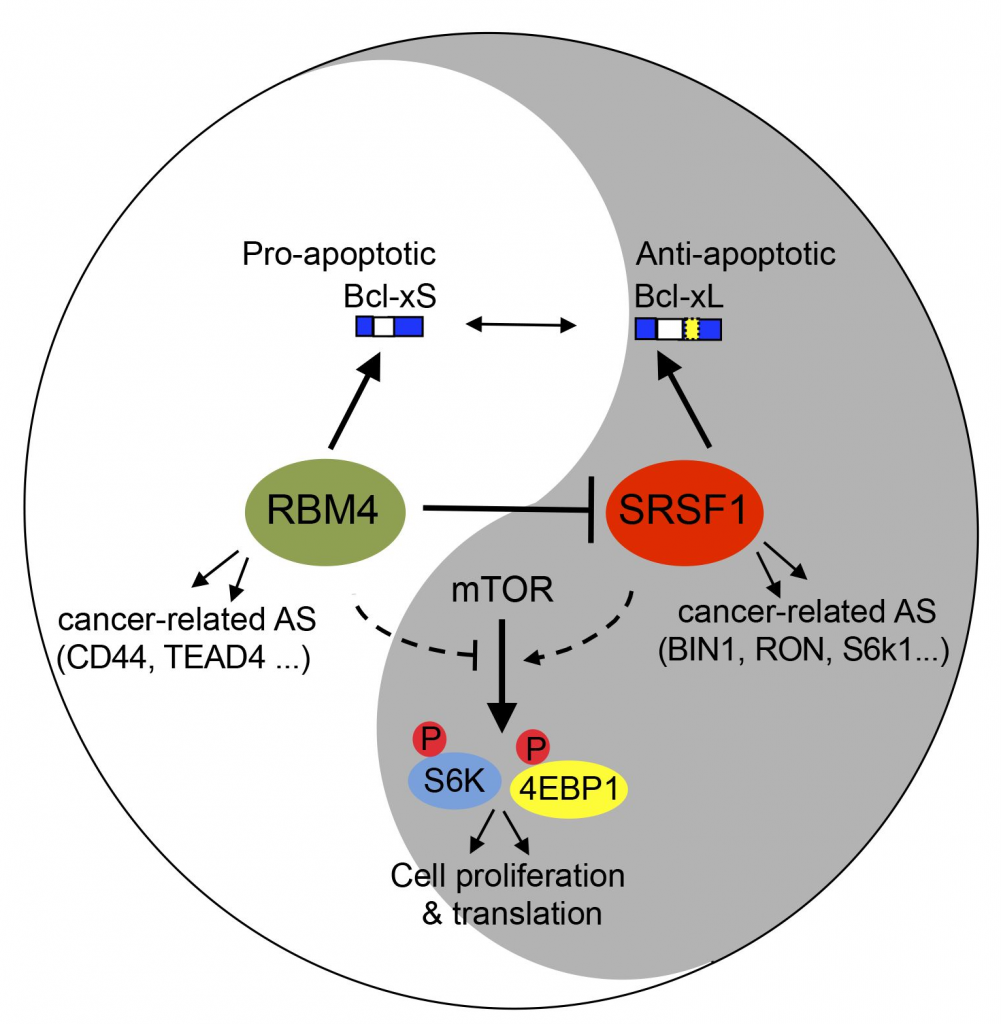
The extensive alteration of AS has been considered to be one of the molecular hallmarks of cancer and is found in numerous genes that affect critical molecular pathways in tumor pathogenesis and progression (e.g. apoptosis, angiogenesis, tumor metastasis). While most of genetic mutations occur at a low frequency in cancer patients (with a few exceptions like TP53), many identified cancer-specific AS events were found in more than half of the tumor samples, suggesting a predominant role of splicing dysregulation in cancer. For example, CD44 is a key mediator of cell-cell and cell-matrix interactions, migration and invasion, and different splicing isoforms of CD44 have been linked with tumor evasion and metastasis in many cancers.
While the global change of splicing in cancers is being increasingly appreciated, the regulatory mechanisms and functional consequences of cancer-specific AS remain poorly understood. Many splicing factors have found to be involved in cancer pathogenesis through mediating AS of hundreds of genes, and they can function as oncogene or tumor suppressor to control splicing of genes involved in different aspects cancer progression. Our lab is aiming to dissect the mechanisms and functional consequence of splicing misregulation in cancer using the combination of molecular biology approaches and genomic approaches.