Projects
- Basic Information
- >> Group Head: Zhu Xinguang
- >> Research Area: Plant Systems Biology
- >> Group Secretary: Wang Sisi
- >> Contact Email: wangsisi0825@163.com

Projects
The ePlant project
Although photosynthesis research has a history of more than a hundred years, it is the astonishing progress in the last two decades that makes this the most well understood process in plants. Even so, our understanding of photosynthesis as a component of the integrated plant system is still quite limited. Specifically, although we have detailed structural, biophysical, biochemical and physiological knowledge of the individual enzymes and enzyme complexes of photosynthesis, we nevertheless lack the understanding and insight into what limits the rate and efficiency of the overall process. As a result, within the intricate complexity of photosynthesis we are unable to effectively pinpoint targets to engineer higher photosynthetic efficiency, even though we have the capacity to increase or decrease the concentration of nearly all the photosynthetic enzymes and functional complexes in photosynthesis. Given the complexity of photosynthesis, the traditional experimental approach, i.e., testing the effects of different combinations of enzyme activities on photosynthetic efficiency, is an unrealistic and insufficient approach. In contrast, employing the deep knowledge of the photosynthetic process to systems integration, and using a quantitative approach based on mathematical models, this provides an efficient means to tackle such issues, as has been demonstrated by the indispensible role of systems modeling and design in the current integrative circuit industry. The overall goal of the ePlant project is to develop mechanistic models of plant primary metabolism (photosynthesis, respiration, nitrogen uptake and assimilation, nitrogen metabolism, and water movements through the plants from the soil to root, stem, leaf until its release into the atmosphere) and its regulation, and integrate these individual models into a functioning systems model (Figure 1). Once built, the e-Plant model will be used as a research platform, firstly, to study the systems properties of plant metabolic and regulatory processes, secondly, to study the molecular mechanisms underlining the adaptation and evolution of plant traits (genetic, metabolic and structural properties related to productivity) under different environments, and thirdly, to identify new ways to engineer higher plant productivity. The current focus of the ePlant project is to develop a mechanistic model of leaf primary metabolism, including photosynthesis and respiration, and incorporate such a leaf level model into a canopy photosynthesis model. Such a model will enable the evaluation of the impacts of leaf level genetic manipulations on total canopy photosynthesis
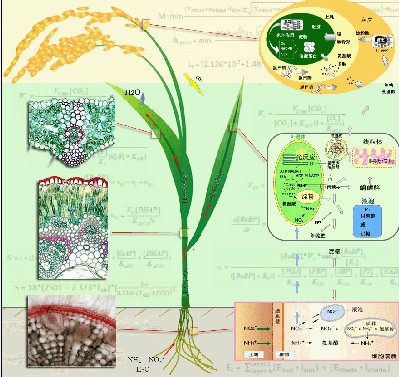
Figure 1. The ePlant model schematic. The overall ePlant model will not only model plant primary metabolism (photosynthesis, respiration, nitrogen assimilation and H2O uptake and transport processes), but also the major regulatory processes and the transport of material among different organs. ePlant will play a key role in guiding the engineering of both food and energy crops to increase their productivity. The ePlant project is an internationally collaborative project with participants ranging from mathematicians, computer scientists, plant biologists and crop breeders. Our group’s primary responsibility is the development of the plant primary metabolism
C4 rice project
With the exception of cool temperature conditions, C4 photosynthesis has a higher photosynthetic energy conversion efficiency compared to C3 photosynthesis. Since the discovery of C4 photosynthesis in the 1960s, significant amounts of research have been conducted into the study of the biochemistry, development and genetics of C4 photosynthesis. Biochemically, the major difference between C3 and C4 photosynthesis is that C4 photosynthesis has a CO2 concentrating mechanism, whichincreases the CO2 concentration around Rubisco and correspondingly suppresses photorespiration (i.e., oxygenation) and increases the net CO2 fixation rate (A). The CO2 concentrating mechanism depends on the cooperation of two specialized cell types, the bundle sheath (BS) and mesophyll (M) cells. In BS cells, the concentration of Rubisco is high, while PSII activity is low; whereas in M cells, the concentration of PEP carboxylase (PEPC), PSII and PSI activities are all maintained at high levels, and Rubisco activity is absent (Sage 2004). Besides the differences in biochemistry between these two cell types, C4 plants have also evolved an efficient metabolite transportation system between the two cell types (Leegood 1999); furthermore, the cell wall of bundle sheath cells are thickened to form a Kranz structure. Numerous lines of evidences suggest that engineering the C4 pathway into C3 crops is feasible, for example, certain tissues in C3 tobacco plants conduct C4-type photosynthesis, and Eleocharis vivipara is able to switch between C3 and C4 photosynthesis dependent on environments (See review in Zhu et al. 2010c). Converting C3 photosynthesis in the major staple crop, e.g. rice, is a feasible approach to dramatically increase crop yield, and correspondingly contribute to alleviating the poverty and hunger of millions in food insecure parts of the world. In addition, enhanced conversion to C4 would significantly improve nitrogen and water use efficiencies. To reach this goal, the Bill & Melinda Gates Foundation (BMBF) has funded a major C4 rice project, led by the International Rice Research Institute, with the ultimate goal of “supercharging” rice with the C4 photosynthetic machinery (Sage andZhu 2011). This BMGF funded C4 rice project has assembled a multidisciplinary team composed of top researchers with expertise in molecular biology, genetics, physiology, biochemistry and mathematics (Figure 2). This team was designed to accelerate progress in understanding C4 photosynthesis, and to develop new techniques and targets for genetic engineering.

Figure 2. Members of the C4 rice consortium. These members are funded by the Bill & Melinda Gates Foundation to engineer the C4 photosynthetic mechanism into rice, which holds potential to increase rice yields up to 40%. Dr. Xinguang Zhu is leading the systems biology and bioinformatics group of the C4 rice project. Other major members of this section include Prof. Tom Brutnell from Boyce Thompson Institute, Prof. Chris Myers from Cornell University and Dr. Richard Bruskiewich from IRRI (Courtesy of Paul Quick).
