Computational Systems Biology Analysis of Single-Cell data
We are developing novel computational and statistical methods to help analyse and better interpret single-cell omic data, with a particular focus on systems biological aspects.
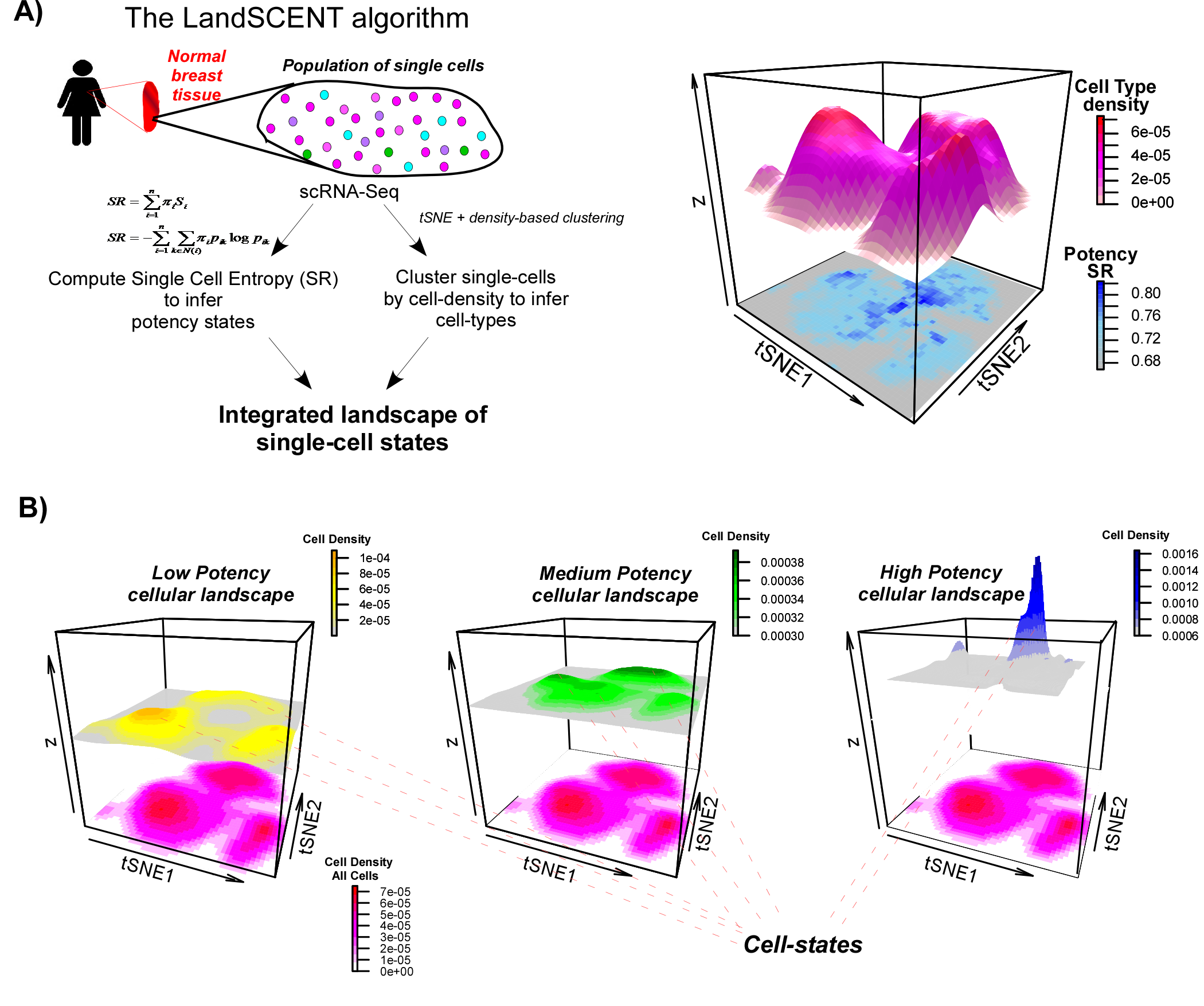
We are developing novel computational and statistical methods to help analyse and better interpret single-cell omic data, with a particular focus on systems biological aspects.

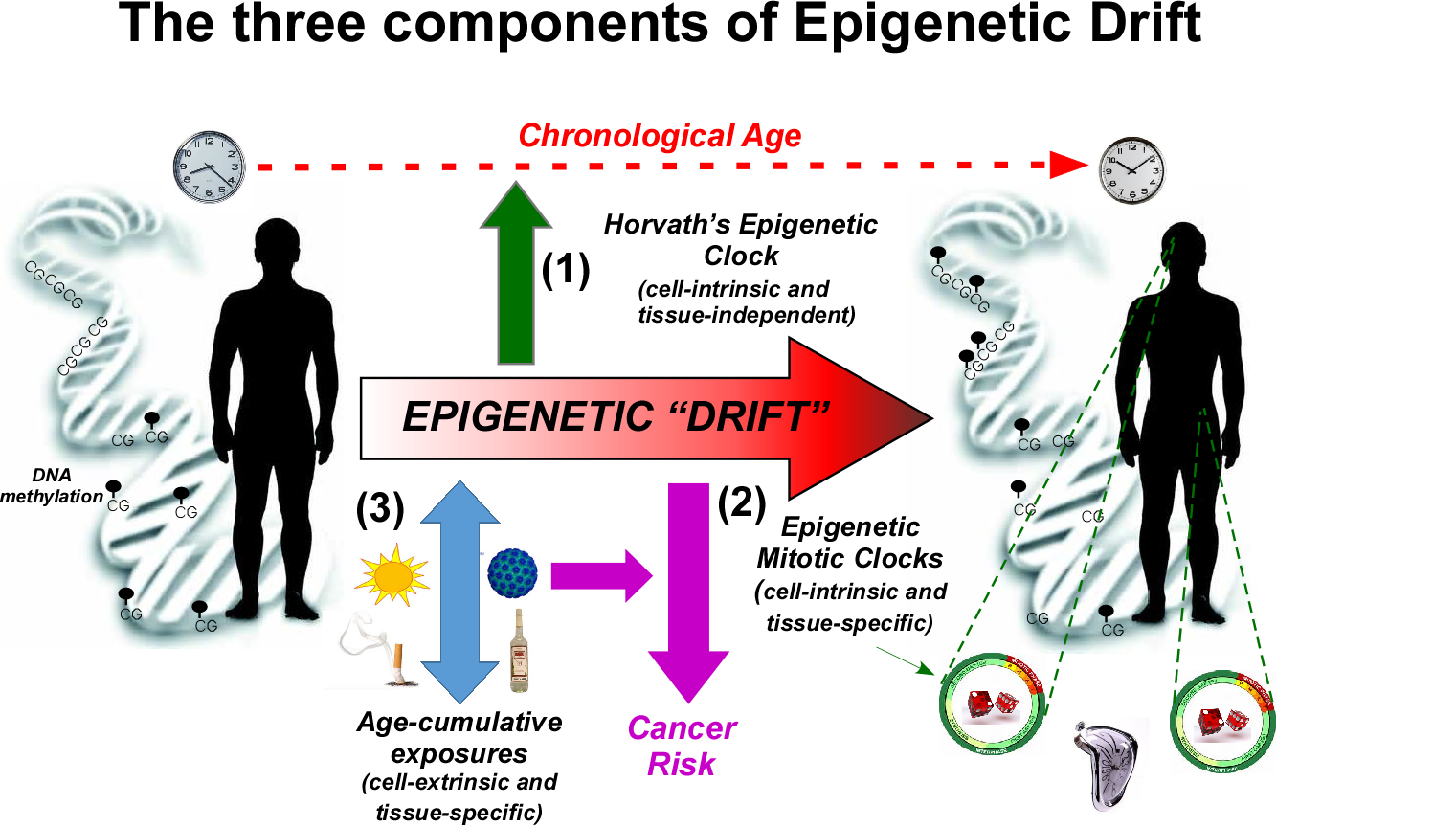
We are currently involved in a large number of data-driven projects concerning the epigenomics of ageing and cancer, in close collaboration with biologists and clinicians at UCL, London, and elsewhere. These include:
(i) Epigenetic stem cell models of cancer: we previously showed that (1) genes, which are required for differentiation of stem cells and which exhibit promoter hypermethylation in cancer, also exhibit age-associated hypermethylation in normal tissue (Teschendorff AE et al Genome Res. 2010, 20(4)), and (2) that these epigenetic changes in normal tissue can be used for cancer risk prediction in the context of cervical cancer (Teschendorff AE et al Genome Med. 2012, 4(3):24.) We are continuing to explore if DNA methylation signatures can indicate the risk of neoplastic transformation in a number of large prospective studies.
(ii) Statistical models of risk prediction: we are particularly interested in developing novel statistical methods for risk prediction and/or early detection, based on epigenomic data collected as part of prospective studies. See Teschendorff AE et al Bioinformatics 2012 Jun1;28(11):1487-94.
(iii) Prognostic biomarkers: we are involved in a number of cancer epigenomic projects, with a focus on women’s cancer. For instance, we recently showed that CpGs that are normally methylated in human embryonic stem cells and which are observed to undergo hypomethylation in cancer, define an epigenetic instability signature indicative of invasion and poor prognosis across all major women cancers. See e.g. Zhuang J et al PloS Genetics 2012 8(2).
(iv) Systems biology of ageing: we have recently shown that epigenetic drift, the gradual deregulation of normal DNA methylation patterns that occurs with age, targets genes exhibiting a very different topology in human interaction networks, compared to other gene-classes associated with ageing and disease (see West J et al PNAS 2013). We are particularly interested in attempting to elucidate the systems biology and functional effects of epigenetic drift.
We are exploring and developing statistical cell-type deconvolution methods to dissect cell-type heterogeneity aimed at epigenome studies performed in general complex tissues.
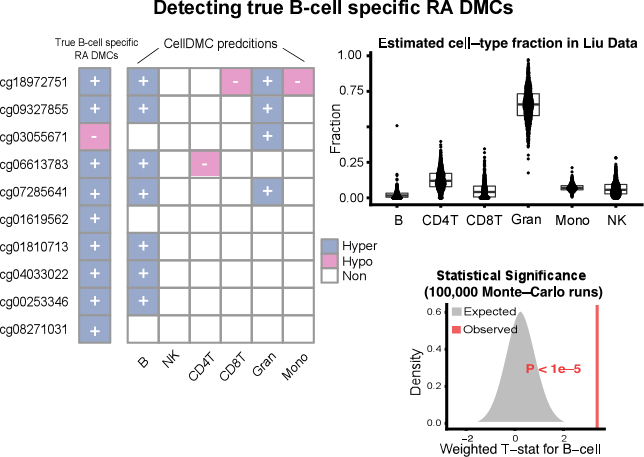
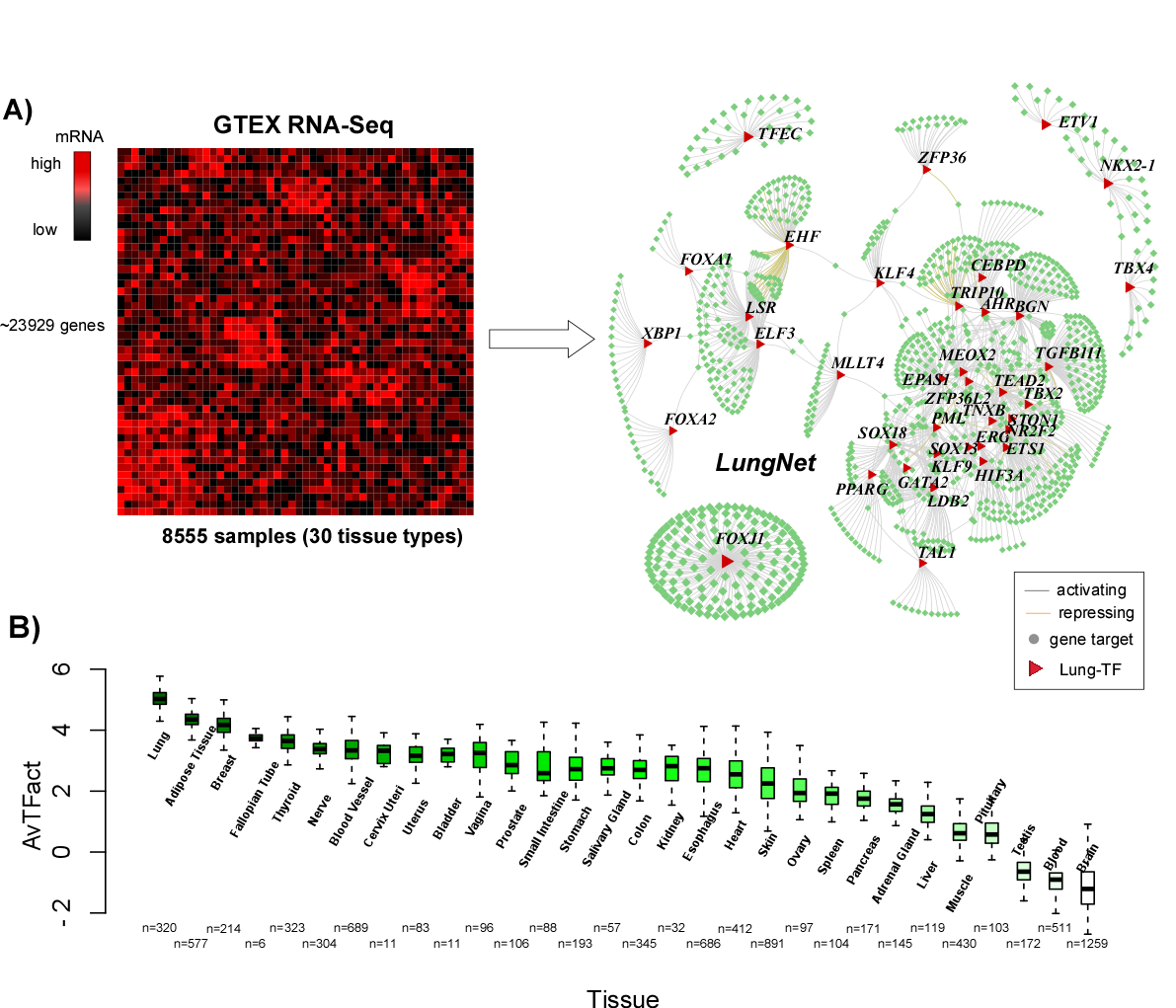
Recent experimental and theoretical work have shown that differential networks are key to understanding the rewiring of the cellular network under endogenous and exogenous perturbations, including those characterizing cancer. We are thus exploring novel tools and concepts from graph theory and network physics to help us elucidate the system-omic principles underlying the cancer phenotype. For example, we recently showed that cancers are characterised by an increased dynamical network entropy and that changes in local network entropy can be used to pinpoint nodes in the network where rewiring takes place (see West J et al Sci Rep 2012 2:802 , Teschendorff AE and Severini S, BMC Systems Biology 2010 4:104). More recently, we are also developing a statistical mechanical framework to understanding basic features of stem cell and developmental biology, such as pluripotency and differentiation.
We are also interested in graph theoretical methods as a means of integrating structural information, such as protein interaction and transcription factor regulatory networks, with multi-dimensional cancer genomic data (mRNA expression, copy number and DNA methylation). The specific goals are to
(i) Identify the genomic/epigenomic aberrations and associated signaling pathways driving differentiation processes and cancer.
(ii) To explore the statistical properties of integrated omic networks and investigate the relationship of these network properties with cancer and other disease phenotypes.
Designed and Coded by Yuan Tian